import rasterio
import numpy as np
import matplotlib.pyplot as plt
from rasterio.plot import show
print(f"Rasterio version: {rasterio.__version__}")Rasterio version: 1.4.3Reading and processing satellite imagery with Python
Rasterio is the essential Python library for reading and writing geospatial raster data.
Since we may not have actual satellite imagery files, let’s create some sample raster data:
# Create sample raster data
def create_sample_raster():
# Create a simple 100x100 raster with some pattern
height, width = 100, 100
# Create coordinate grids
x = np.linspace(-2, 2, width)
y = np.linspace(-2, 2, height)
X, Y = np.meshgrid(x, y)
# Create a pattern (e.g., circular pattern)
data = np.sin(np.sqrt(X**2 + Y**2)) * 255
data = data.astype(np.uint8)
return data
sample_data = create_sample_raster()
print(f"Sample data shape: {sample_data.shape}")
print(f"Data type: {sample_data.dtype}")
print(f"Data range: {sample_data.min()} to {sample_data.max()}")Sample data shape: (100, 100)
Data type: uint8
Data range: 7 to 254# Basic statistics
print(f"Mean: {np.mean(sample_data):.2f}")
print(f"Standard deviation: {np.std(sample_data):.2f}")
print(f"Unique values: {len(np.unique(sample_data))}")
# Histogram of values
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.imshow(sample_data, cmap='viridis')
plt.title('Sample Raster Data')
plt.colorbar()
plt.subplot(1, 2, 2)
plt.hist(sample_data.flatten(), bins=50, alpha=0.7)
plt.title('Histogram of Values')
plt.xlabel('Pixel Value')
plt.ylabel('Frequency')
plt.tight_layout()
plt.show()Mean: 215.01
Standard deviation: 46.13
Unique values: 182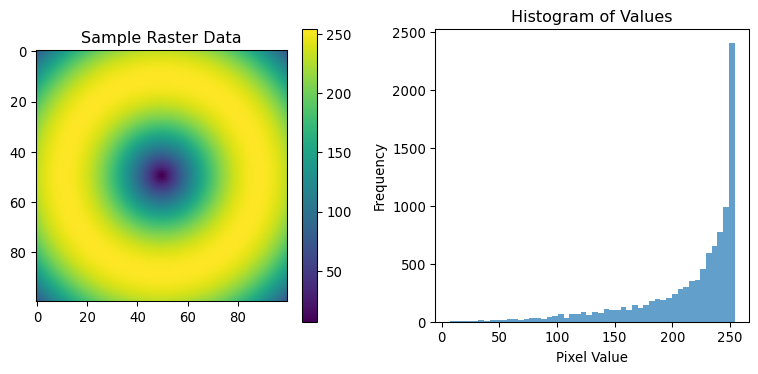
# Create masks
high_values = sample_data > 200
low_values = sample_data < 50
print(f"Pixels with high values (>200): {np.sum(high_values)}")
print(f"Pixels with low values (<50): {np.sum(low_values)}")
# Apply mask
masked_data = np.where(high_values, sample_data, 0)
plt.figure(figsize=(12, 4))
plt.subplot(1, 3, 1)
plt.imshow(sample_data, cmap='viridis')
plt.title('Original Data')
plt.subplot(1, 3, 2)
plt.imshow(high_values, cmap='gray')
plt.title('High Value Mask')
plt.subplot(1, 3, 3)
plt.imshow(masked_data, cmap='viridis')
plt.title('Masked Data')
plt.tight_layout()
plt.show()Pixels with high values (>200): 7364
Pixels with low values (<50): 76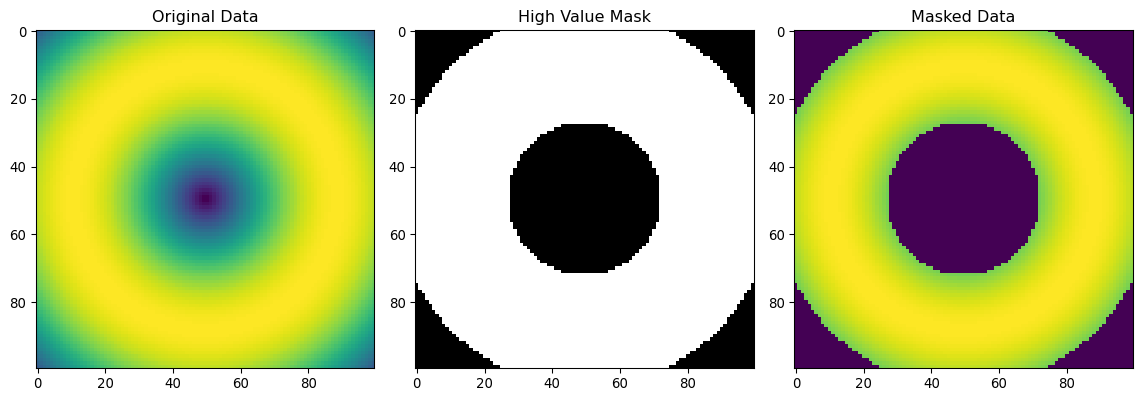
from rasterio.transform import from_bounds
# Create a sample transform (geographic coordinates)
west, south, east, north = -120.0, 35.0, -115.0, 40.0 # Bounding box
transform = from_bounds(west, south, east, north, 100, 100)
print(f"Transform: {transform}")
print(f"Pixel width: {transform[0]}")
print(f"Pixel height: {abs(transform[4])}")
# Convert pixel coordinates to geographic coordinates
def pixel_to_geo(row, col, transform):
x, y = rasterio.transform.xy(transform, row, col)
return x, y
# Example: center pixel
center_row, center_col = 50, 50
geo_x, geo_y = pixel_to_geo(center_row, center_col, transform)
print(f"Center pixel ({center_row}, {center_col}) -> Geographic: ({geo_x:.2f}, {geo_y:.2f})")Transform: | 0.05, 0.00,-120.00|
| 0.00,-0.05, 40.00|
| 0.00, 0.00, 1.00|
Pixel width: 0.05
Pixel height: 0.05
Center pixel (50, 50) -> Geographic: (-117.47, 37.48)# Create a profile for writing raster data
profile = {
'driver': 'GTiff',
'dtype': 'uint8',
'nodata': None,
'width': 100,
'height': 100,
'count': 1,
'crs': 'EPSG:4326', # WGS84 lat/lon
'transform': transform
}
print("Raster profile:")
for key, value in profile.items():
print(f" {key}: {value}")Raster profile:
driver: GTiff
dtype: uint8
nodata: None
width: 100
height: 100
count: 1
crs: EPSG:4326
transform: | 0.05, 0.00,-120.00|
| 0.00,-0.05, 40.00|
| 0.00, 0.00, 1.00|# Create multi-band sample data (RGB-like)
def create_multiband_sample():
height, width = 100, 100
bands = 3
# Create different patterns for each band
x = np.linspace(-2, 2, width)
y = np.linspace(-2, 2, height)
X, Y = np.meshgrid(x, y)
# Band 1: Circular pattern
band1 = (np.sin(np.sqrt(X**2 + Y**2)) * 127 + 127).astype(np.uint8)
# Band 2: Linear gradient
band2 = (np.linspace(0, 255, width) * np.ones((height, 1))).astype(np.uint8)
# Band 3: Checkerboard pattern
band3 = ((X + Y) > 0).astype(np.uint8) * 255
return np.stack([band1, band2, band3])
multiband_data = create_multiband_sample()
print(f"Multiband shape: {multiband_data.shape}")
print(f"Shape format: (bands, height, width)")Multiband shape: (3, 100, 100)
Shape format: (bands, height, width)fig, axes = plt.subplots(2, 2, figsize=(10, 10))
# Individual bands
for i in range(3):
row, col = i // 2, i % 2
axes[row, col].imshow(multiband_data[i], cmap='gray')
axes[row, col].set_title(f'Band {i+1}')
# RGB composite (transpose for matplotlib)
rgb_composite = np.transpose(multiband_data, (1, 2, 0))
axes[1, 1].imshow(rgb_composite)
axes[1, 1].set_title('RGB Composite')
plt.tight_layout()
plt.show()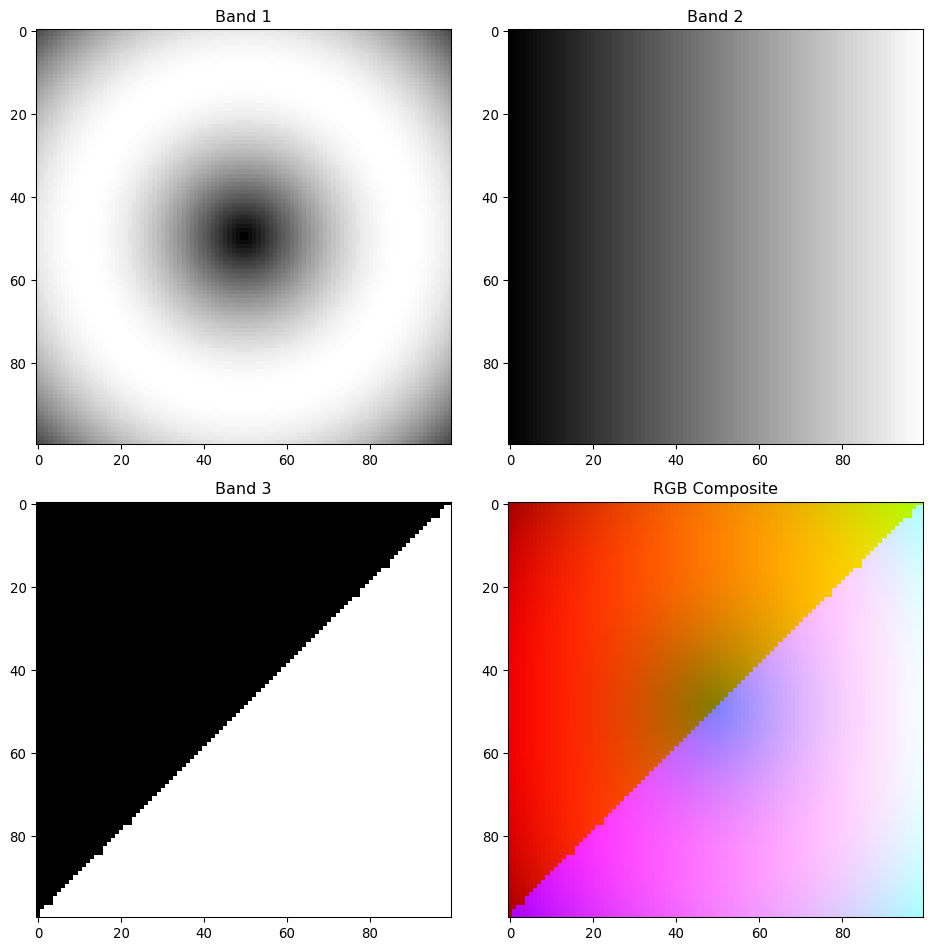
# Simulate NIR and Red bands
nir_band = multiband_data[0].astype(np.float32)
red_band = multiband_data[1].astype(np.float32)
# Calculate NDVI-like index
# NDVI = (NIR - Red) / (NIR + Red)
ndvi = (nir_band - red_band) / (nir_band + red_band + 1e-8) # Small value to avoid division by zero
plt.figure(figsize=(12, 4))
plt.subplot(1, 3, 1)
plt.imshow(nir_band, cmap='RdYlBu_r')
plt.title('NIR-like Band')
plt.colorbar()
plt.subplot(1, 3, 2)
plt.imshow(red_band, cmap='Reds')
plt.title('Red-like Band')
plt.colorbar()
plt.subplot(1, 3, 3)
plt.imshow(ndvi, cmap='RdYlGn', vmin=-1, vmax=1)
plt.title('NDVI-like Index')
plt.colorbar()
plt.tight_layout()
plt.show()
print(f"NDVI range: {ndvi.min():.3f} to {ndvi.max():.3f}")
print(f"NDVI mean: {np.mean(ndvi):.3f}")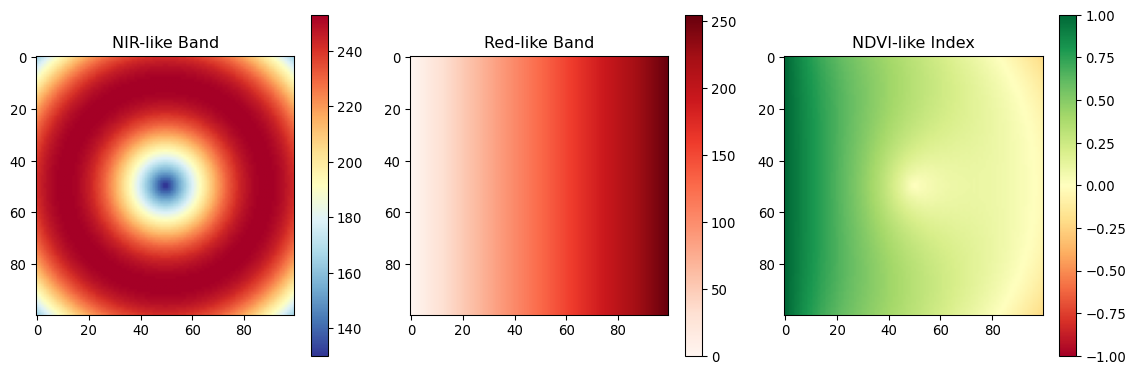
NDVI range: -0.211 to 1.000
NDVI mean: 0.353from scipy import ndimage
# Demonstrate different resampling methods
original = sample_data
# Downsample (reduce resolution)
downsampled = ndimage.zoom(original, 0.5, order=1) # Linear interpolation
# Upsample (increase resolution)
upsampled = ndimage.zoom(original, 2.0, order=1) # Linear interpolation
plt.figure(figsize=(15, 4))
plt.subplot(1, 3, 1)
plt.imshow(original, cmap='viridis')
plt.title(f'Original ({original.shape[0]}x{original.shape[1]})')
plt.subplot(1, 3, 2)
plt.imshow(downsampled, cmap='viridis')
plt.title(f'Downsampled ({downsampled.shape[0]}x{downsampled.shape[1]})')
plt.subplot(1, 3, 3)
plt.imshow(upsampled, cmap='viridis')
plt.title(f'Upsampled ({upsampled.shape[0]}x{upsampled.shape[1]})')
plt.tight_layout()
plt.show()
print(f"Original size: {original.shape}")
print(f"Downsampled size: {downsampled.shape}")
print(f"Upsampled size: {upsampled.shape}")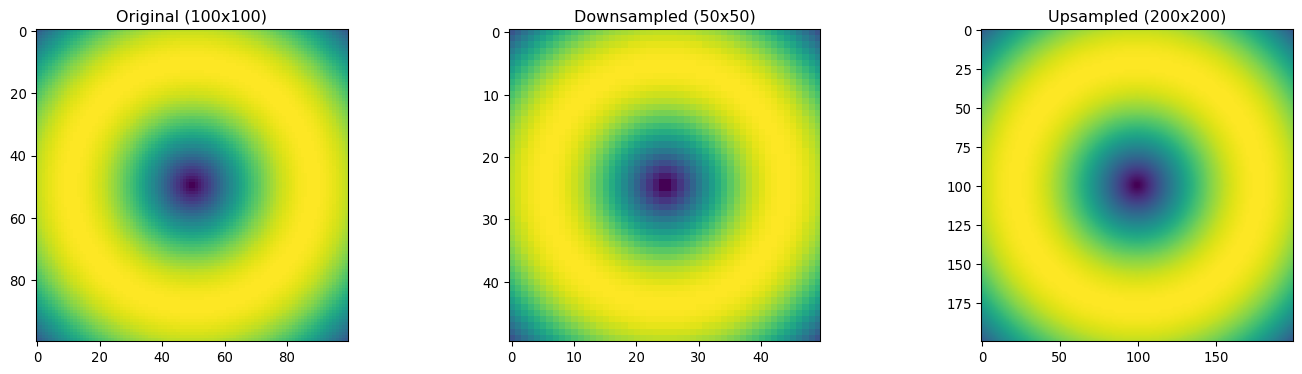
Original size: (100, 100)
Downsampled size: (50, 50)
Upsampled size: (200, 200)# Simulate processing large rasters in blocks
def process_in_blocks(data, block_size=50):
"""Process data in blocks to simulate handling large rasters"""
height, width = data.shape
processed = np.zeros_like(data)
for row in range(0, height, block_size):
for col in range(0, width, block_size):
# Define window bounds
row_end = min(row + block_size, height)
col_end = min(col + block_size, width)
# Extract block
block = data[row:row_end, col:col_end]
# Process block (example: enhance contrast)
processed_block = np.clip(block * 1.2, 0, 255)
# Write back to result
processed[row:row_end, col:col_end] = processed_block
print(f"Processed block: ({row}:{row_end}, {col}:{col_end})")
return processed
# Process sample data in blocks
processed_data = process_in_blocks(sample_data, block_size=25)
plt.figure(figsize=(10, 4))
plt.subplot(1, 2, 1)
plt.imshow(sample_data, cmap='viridis')
plt.title('Original')
plt.subplot(1, 2, 2)
plt.imshow(processed_data, cmap='viridis')
plt.title('Processed (Enhanced)')
plt.tight_layout()
plt.show()Processed block: (0:25, 0:25)
Processed block: (0:25, 25:50)
Processed block: (0:25, 50:75)
Processed block: (0:25, 75:100)
Processed block: (25:50, 0:25)
Processed block: (25:50, 25:50)
Processed block: (25:50, 50:75)
Processed block: (25:50, 75:100)
Processed block: (50:75, 0:25)
Processed block: (50:75, 25:50)
Processed block: (50:75, 50:75)
Processed block: (50:75, 75:100)
Processed block: (75:100, 0:25)
Processed block: (75:100, 25:50)
Processed block: (75:100, 50:75)
Processed block: (75:100, 75:100)
Key Rasterio concepts covered: - Reading and understanding raster data structure - Working with single and multi-band imagery - Coordinate transforms and geospatial metadata
- Band math and index calculations - Resampling and processing techniques - Block-based processing for large datasets
---
title: "Working with Rasterio"
subtitle: "Reading and processing satellite imagery with Python"
jupyter: geoai
format:
html:
code-fold: false
---
## Introduction to Rasterio
Rasterio is the essential Python library for reading and writing geospatial raster data.
```{python}
import rasterio
import numpy as np
import matplotlib.pyplot as plt
from rasterio.plot import show
print(f"Rasterio version: {rasterio.__version__}")
```
## Creating Sample Data
Since we may not have actual satellite imagery files, let's create some sample raster data:
```{python}
# Create sample raster data
def create_sample_raster():
# Create a simple 100x100 raster with some pattern
height, width = 100, 100
# Create coordinate grids
x = np.linspace(-2, 2, width)
y = np.linspace(-2, 2, height)
X, Y = np.meshgrid(x, y)
# Create a pattern (e.g., circular pattern)
data = np.sin(np.sqrt(X**2 + Y**2)) * 255
data = data.astype(np.uint8)
return data
sample_data = create_sample_raster()
print(f"Sample data shape: {sample_data.shape}")
print(f"Data type: {sample_data.dtype}")
print(f"Data range: {sample_data.min()} to {sample_data.max()}")
```
## Working with Raster Arrays
### Basic array operations
```{python}
# Basic statistics
print(f"Mean: {np.mean(sample_data):.2f}")
print(f"Standard deviation: {np.std(sample_data):.2f}")
print(f"Unique values: {len(np.unique(sample_data))}")
# Histogram of values
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.imshow(sample_data, cmap='viridis')
plt.title('Sample Raster Data')
plt.colorbar()
plt.subplot(1, 2, 2)
plt.hist(sample_data.flatten(), bins=50, alpha=0.7)
plt.title('Histogram of Values')
plt.xlabel('Pixel Value')
plt.ylabel('Frequency')
plt.tight_layout()
plt.show()
```
### Array masking and filtering
```{python}
# Create masks
high_values = sample_data > 200
low_values = sample_data < 50
print(f"Pixels with high values (>200): {np.sum(high_values)}")
print(f"Pixels with low values (<50): {np.sum(low_values)}")
# Apply mask
masked_data = np.where(high_values, sample_data, 0)
plt.figure(figsize=(12, 4))
plt.subplot(1, 3, 1)
plt.imshow(sample_data, cmap='viridis')
plt.title('Original Data')
plt.subplot(1, 3, 2)
plt.imshow(high_values, cmap='gray')
plt.title('High Value Mask')
plt.subplot(1, 3, 3)
plt.imshow(masked_data, cmap='viridis')
plt.title('Masked Data')
plt.tight_layout()
plt.show()
```
## Raster Metadata and Transforms
### Understanding geospatial transforms
```{python}
from rasterio.transform import from_bounds
# Create a sample transform (geographic coordinates)
west, south, east, north = -120.0, 35.0, -115.0, 40.0 # Bounding box
transform = from_bounds(west, south, east, north, 100, 100)
print(f"Transform: {transform}")
print(f"Pixel width: {transform[0]}")
print(f"Pixel height: {abs(transform[4])}")
# Convert pixel coordinates to geographic coordinates
def pixel_to_geo(row, col, transform):
x, y = rasterio.transform.xy(transform, row, col)
return x, y
# Example: center pixel
center_row, center_col = 50, 50
geo_x, geo_y = pixel_to_geo(center_row, center_col, transform)
print(f"Center pixel ({center_row}, {center_col}) -> Geographic: ({geo_x:.2f}, {geo_y:.2f})")
```
### Creating profiles for writing rasters
```{python}
# Create a profile for writing raster data
profile = {
'driver': 'GTiff',
'dtype': 'uint8',
'nodata': None,
'width': 100,
'height': 100,
'count': 1,
'crs': 'EPSG:4326', # WGS84 lat/lon
'transform': transform
}
print("Raster profile:")
for key, value in profile.items():
print(f" {key}: {value}")
```
## Multi-band Operations
### Working with multi-band data
```{python}
# Create multi-band sample data (RGB-like)
def create_multiband_sample():
height, width = 100, 100
bands = 3
# Create different patterns for each band
x = np.linspace(-2, 2, width)
y = np.linspace(-2, 2, height)
X, Y = np.meshgrid(x, y)
# Band 1: Circular pattern
band1 = (np.sin(np.sqrt(X**2 + Y**2)) * 127 + 127).astype(np.uint8)
# Band 2: Linear gradient
band2 = (np.linspace(0, 255, width) * np.ones((height, 1))).astype(np.uint8)
# Band 3: Checkerboard pattern
band3 = ((X + Y) > 0).astype(np.uint8) * 255
return np.stack([band1, band2, band3])
multiband_data = create_multiband_sample()
print(f"Multiband shape: {multiband_data.shape}")
print(f"Shape format: (bands, height, width)")
```
### Visualizing multi-band data
```{python}
fig, axes = plt.subplots(2, 2, figsize=(10, 10))
# Individual bands
for i in range(3):
row, col = i // 2, i % 2
axes[row, col].imshow(multiband_data[i], cmap='gray')
axes[row, col].set_title(f'Band {i+1}')
# RGB composite (transpose for matplotlib)
rgb_composite = np.transpose(multiband_data, (1, 2, 0))
axes[1, 1].imshow(rgb_composite)
axes[1, 1].set_title('RGB Composite')
plt.tight_layout()
plt.show()
```
## Band Math and Indices
### Calculate vegetation index (NDVI-like)
```{python}
# Simulate NIR and Red bands
nir_band = multiband_data[0].astype(np.float32)
red_band = multiband_data[1].astype(np.float32)
# Calculate NDVI-like index
# NDVI = (NIR - Red) / (NIR + Red)
ndvi = (nir_band - red_band) / (nir_band + red_band + 1e-8) # Small value to avoid division by zero
plt.figure(figsize=(12, 4))
plt.subplot(1, 3, 1)
plt.imshow(nir_band, cmap='RdYlBu_r')
plt.title('NIR-like Band')
plt.colorbar()
plt.subplot(1, 3, 2)
plt.imshow(red_band, cmap='Reds')
plt.title('Red-like Band')
plt.colorbar()
plt.subplot(1, 3, 3)
plt.imshow(ndvi, cmap='RdYlGn', vmin=-1, vmax=1)
plt.title('NDVI-like Index')
plt.colorbar()
plt.tight_layout()
plt.show()
print(f"NDVI range: {ndvi.min():.3f} to {ndvi.max():.3f}")
print(f"NDVI mean: {np.mean(ndvi):.3f}")
```
## Resampling and Reprojection Concepts
### Understanding resampling methods
```{python}
from scipy import ndimage
# Demonstrate different resampling methods
original = sample_data
# Downsample (reduce resolution)
downsampled = ndimage.zoom(original, 0.5, order=1) # Linear interpolation
# Upsample (increase resolution)
upsampled = ndimage.zoom(original, 2.0, order=1) # Linear interpolation
plt.figure(figsize=(15, 4))
plt.subplot(1, 3, 1)
plt.imshow(original, cmap='viridis')
plt.title(f'Original ({original.shape[0]}x{original.shape[1]})')
plt.subplot(1, 3, 2)
plt.imshow(downsampled, cmap='viridis')
plt.title(f'Downsampled ({downsampled.shape[0]}x{downsampled.shape[1]})')
plt.subplot(1, 3, 3)
plt.imshow(upsampled, cmap='viridis')
plt.title(f'Upsampled ({upsampled.shape[0]}x{upsampled.shape[1]})')
plt.tight_layout()
plt.show()
print(f"Original size: {original.shape}")
print(f"Downsampled size: {downsampled.shape}")
print(f"Upsampled size: {upsampled.shape}")
```
## Common Rasterio Patterns
### Working with windows and blocks
```{python}
# Simulate processing large rasters in blocks
def process_in_blocks(data, block_size=50):
"""Process data in blocks to simulate handling large rasters"""
height, width = data.shape
processed = np.zeros_like(data)
for row in range(0, height, block_size):
for col in range(0, width, block_size):
# Define window bounds
row_end = min(row + block_size, height)
col_end = min(col + block_size, width)
# Extract block
block = data[row:row_end, col:col_end]
# Process block (example: enhance contrast)
processed_block = np.clip(block * 1.2, 0, 255)
# Write back to result
processed[row:row_end, col:col_end] = processed_block
print(f"Processed block: ({row}:{row_end}, {col}:{col_end})")
return processed
# Process sample data in blocks
processed_data = process_in_blocks(sample_data, block_size=25)
plt.figure(figsize=(10, 4))
plt.subplot(1, 2, 1)
plt.imshow(sample_data, cmap='viridis')
plt.title('Original')
plt.subplot(1, 2, 2)
plt.imshow(processed_data, cmap='viridis')
plt.title('Processed (Enhanced)')
plt.tight_layout()
plt.show()
```
## Summary
Key Rasterio concepts covered:
- Reading and understanding raster data structure
- Working with single and multi-band imagery
- Coordinate transforms and geospatial metadata
- Band math and index calculations
- Resampling and processing techniques
- Block-based processing for large datasets